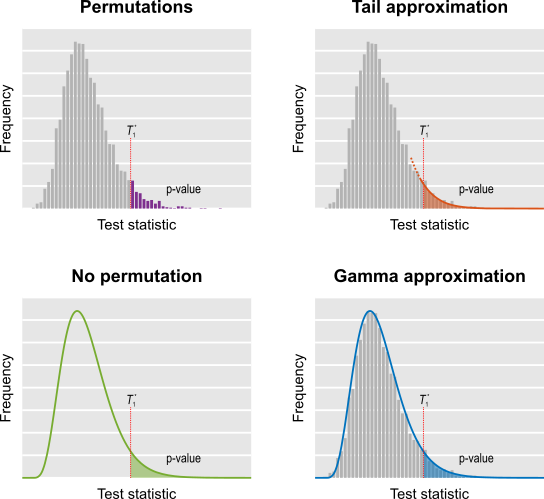
In brain imaging, each voxel (or vertex, or face, or edge) constitutes a single statistical test. Because thousands such voxels are present in an image, a single experiment results in thousands of statistical tests being performed. The p-value is the probability of finding a test statistic at least as large as the one observed in a given voxel, provided that no effect is present. A p-value of 0.05 indicates that, if an experiment is repeated 20 times and there are no effects, on average one of these repetitions will be considered significant. If thousands of tests are performed, the chance of obtaining a spuriously significant result in at least one voxel increases: if there are 1000 voxels, and at the same test level , we expect, on average, to find 50 significant tests, even in the absence of any effect. This is known as the multiple testing problem. A review of the topic for brain imaging provided in Nichols and Hayasaka (2003) [see references at the end].
To take the multiple testing problem into account, either the test level (), or the p-values can be adjusted, such that instead of controlling the error rate at each individual test, the error rate is controlled for the whole set (family) of tests. Controlling such family-wise error rate (FWER) ensures that the chance of finding a significant result anywhere in the image is expected to be within a certain predefined level. For example, if there are 1000 voxels, and the FWER-adjusted test level is 0.05, we expect that, if the experiment is repeated for all the voxels 20 times, then on average in one of these repetitions there will be an error somewhere in the image. The adjustment of the p-values or of the test level is done using the distribution of the maximum statistic, something that most readers of this blog are certainly well aware of, as that permeates most of the imaging literature since the early 1990s.
Have you ever wondered why? What is so special about the distribution of the maximum that makes it useful to correct the error rate when there are multiple tests?
Definitions first
Say we have a set of voxels in an image. For a given voxel
,
, with test statistic
, the probability that
is larger than some cutoff
is denoted by:
where is the cumulative distribution function (cdf) of the test statistic. If the cutoff
is used to accept or reject a hypothesis, then we say that we have a false positive if an observed
is larger than
when there is no actual true effect. A false positive is also known as error type I (in this post, the only type of error discussed is of the type I).
For an image (or any other set of tests), if there is an error anywhere, we say that a family-wise error has occurred. We can therefore define a “family-wise null hypothesis” that there is no signal anywhere; to reject this hypothesis, it suffices to have a single, lonely voxel in which . With many voxels, the chances of this happening increase, even if there no effect is present. We can, however, adjust our cuttoff
to some other value
so that the probability of rejecting such family-wise null hypothesis remains within a certain level, say
.
Union-intersection tests
The “family-wise null hypothesis” is effectively a joint null hypothesis that there is no effect anywhere. That is, it is an union-intersection test (UIT; Roy, 1953). This joint hypothesis is retained if all tests have statistics that are below the significance cutoff. What is the probability of this happening? From the above we know that . The probability of the same happening for all voxels simultaneously is, therefore, simply the product of such probabilities, assuming of course that the voxels are all independent:
Thus, the probability that any voxel has a significant result, which would lead to the occurrence of a family-wise error, is . If all voxels have identical distribution under the null, then the same is stated as
.
Distribution of the maximum
Consider the maximum of the set of voxels, that is,
. The random variable
is only smaller or equal than some cutoff
if all values
are smaller or equal than
. If the voxels are independent, this enables us to derive the cdf of
:
.
Thus, the probability that is larger than some threshold
is
. If all voxels have identical distribution under the null, then the same is stated as
.
These results, lo and behold, are the same as those used for the UIT above, hence how the distribution of the maximum can be used to control the family-wise error rate (if the distribution of the maximum is computed via permutations, independence is not required).
Closure
The above is not the only way in which we can see why the distribution of the maximum allows the control of the family-wise error rate. The work by Marcus, Peritz and Gabriel (1976) showed that, in the context of multiple testing, the null hypothesis for a particular test can be rejected provided that all possible joint (multivariate) tests done within the set and including
are also significant, and doing so controls the family-wise error rate. For example, if there are four tests,
, the test in
is considered significant if the joint tests using (1,2,3,4), (1,2,3), (1,2,4), (1,3,4), (1,2), (1,3), (1,4) and (1) are all significant (that is, all that include
). Such joint test can be quite much any valid test, including Hotelling’s
, MANOVA/MANCOVA, or NPC (Non-Parametric Combination), all of which are based on recomputing the test statistic from the original data, or others, based on the test statistics or p-values of each of the elementary
tests, as in a meta-analysis.
Such closed testing procedure (CTP) incurs an additional problem, though: the number of joint tests that needs to be done is , which in imaging applications renders them unfeasible. However, there is one particular joint test that provides a direct algorithmic shortcut: using the
as the test statistic for the joint test. The maximum across all
tests is also the maximum for any subset of tests, such that these can be skipped altogether. This gives a vastly efficient algorithmic shortcut to a CTP, as shown by Westfall and Young (1993).
Simple intuition
One does not need to chase the original papers cited above (although doing so cannot hurt). Broadly, the same can be concluded based solely on intuition: if the distribution of some test statistic that is not the distribution of the maximum within an image were used as the reference to compute the (FWER-adjusted) p-values at a given voxel , then the probability of finding a voxel with a test statistic larger than
anywhere could not be determined: there could always be some other voxel
, with an even larger statistic (i.e.,
), but the probability of such happening would not be captured by the distribution of a non-maximum. Hence the chance of finding a significant voxel anywhere in the image under the null hypothesis (the very definition of FWER) would not be controlled. Using the absolute maximum eliminates this logical leakage.
References
- Marcus R, Peritz E, Gabriel KR. On closed testing pocedures with special reference to ordered analysis of variance. Biometrika. 1976 Dec;63(3):655.
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res. 2003 Oct;12(5):419–46.
- Roy SN. On a heuristic method of test construction and its use in multivariate analysis. Ann Math Stat. 1953 Jun;24(2):220–38.
- Westfall PH, Young SS. Resampling-based multiple testing: examples and methods for p-value adjustment. New York, Wiley, 1993.