Permutation tests are more robust and help to make scientific results more reproducible by depending on fewer assumptions. However, they are computationally intensive as recomputing a model thousands of times can be slow. The purpose of this post is to briefly list some options available for speeding up permutation.
Firstly, no speed-ups may be needed: for small sample sizes, or low resolutions, or small regions of interest, a permutation test can run in a matter of minutes. For larger data, however, accelerations may be of use. One option is acceleration through parallel processing or GPUs (for example applications of the latter, see Eklund et al., 2012, Eklund et al., 2013 and Hernández et al., 2013; references below), though this does require specialised implementation. Another option is to reduce the computational burden by exploiting the properties of the statistics and their distributions. A menu of options includes:
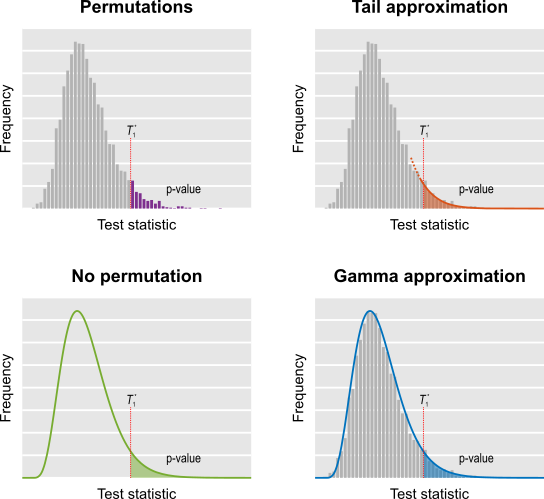
- Do few permutations (shorthand name: fewperms). The results remain valid on average, although the p-values will have higher variability.
- Keep permuting until a fixed number of permutations with statistic larger than the unpermuted is found (a.k.a., negative binomial; shorthand name: negbin).
- Do a few permutations, then approximate the tail of the permutation distribution by fitting a generalised Pareto distribution to its tail (shorthand name: tail).
- Approximate the permutation distribution with a gamma distribution, using simple properties of the test statistic itself, amazingly not requiring any permutations at all (shorthand name: noperm).
- Do a few permutations, then approximate the full permutation distribution by fitting a gamma distribution (shorthand name: gamma).
- Run permutations on only a few voxels, then fill the missing ones using low-rank matrix completion theory (shorthand name: lowrank).
These strategies allow accelerations >100x, yielding nearly identical results as in the non-accelerated case. Some, such as tail approximation, are generic enough to be used nearly all the most common scenarios, including univariate and multivariate tests, spatial statistics, and for correction for multiple testing.
In addition to accelerating permutation tests, some of these strategies, such as tail and noperm, allow continuous p-values to be found, and refine the p-values far into the tail of the distribution, thus avoiding the usual discreteness of p-values, which can be a problem in some applications if too few permutations are done.
These methods are available in the tool PALM — Permutation Analysis of Linear Models — and the complete description, evaluation, and application to the re-analysis of a voxel-based morphometry study (Douaud et al., 2007) have been just published in Winkler et al., 2016 (for the Supplementary Material, click here). The paper includes a flow chart prescribing these various approaches for each case, reproduced below.
The hope is that these accelerations will facilitate the use of permutation tests and, if used in combination with hardware and/or software improvements, can further expedite computation leaving little reason not to use these tests.
References
- Douaud G, Smith S, Jenkinson M, Behrens T, Johansen-Berg H, Vickers J, James S, Voets N, Watkins K, Matthews PM, James A. Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain. 2007 Sep;130(Pt 9):2375-86. Epub 2007 Aug 13.
- Eklund A, Andersson M, Knutsson H. fMRI analysis on the GPU-possibilities and challenges. Comput Methods Programs Biomed. 2012 Feb;105(2):145-61.
- Eklund A, Dufort P, Forsberg D, LaConte SM. Medical image processing on the GPU – Past, present and future. Med Image Anal. 2013;17(8):1073-94.
- Hernández M, Guerrero GD, Cecilia JM, García JM, Inuggi A, Jbabdi S, et al. Accelerating fibre orientation estimation from diffusion weighted magnetic resonance imaging using GPUs. PLoS One. 2013 Jan;8(4):e61892.1.
- Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. Neuroimage. 2014 May 15;92:381-97.
- Winkler AM, Ridgway GR, Douaud G, Nichols TE, Smith SM. Faster permutation inference in brain imaging. Neuroimage. 2016 Jun 7;141:502-516.
Contributed to this post: Tom Nichols, Ged Ridgway.


Silly question: how many MC is a full permutation? you can run simulations to evaluate the minimum needed to have ‘stable results’. We have done that for eeg with Tom, and found cluster inference (mass or tfce) needs at least 800 iterations (http://www.sciencedirect.com/science/article/pii/S0165027014002878). So we do 1000 as default, no need to do more. The same could be estimated for MRI and fMRI (as I suspect is is data type dependent) .
Cyril
Hi Cyril,
I think the suspicion that it’d be data type dependent is a strong one. We can compute the confidence interval and the resampling risk is also an evidence, although the latter depends on the strength of the signal, and that is dataset dependent.
In the paper the “full” is the set of 50k permutations. Some methods allowed undistinguishable results with 500 permutations only.
Cheers,
Anderson
Hi Anderson, speaking of doing things faster :)
I have attempted to run PALM on cluster. At first, to load on cluster, I used qsub with a command, which had been working well when running from the local disk. I modified the “palm” script to change IS_CLUSTER=1 and provided an address for the “tmp” file. The job was run, but there was no feedback from the system and what is more important, no output was generated.
To troubleshoot, I ran palm without qsub and found it was throwing an exception, specifically it was something to the extent of “MATLAB is unable to recognize | “, i.e., a “pipe” character.
Among the suspects is the fact, that we’ve gotten an older 2014 version of MATLAB installed on our cluster as opposed to 2016 on my iMac locally. Could that be a compatibility issue?
Thank you,
~ Sergey
Hi Sergey,
All arguments supplied are passed to MATLAB so that the pipe goes along with it. If you are using pipe, i.e., the symbol “|”, to grep some specific outputs that are printed in the screen, no worries: in most cluster environments all outputs are automatically stored in a log-file, which can be perused later. If there is some other reason to use “|”, please let me know and perhaps there may be some other solution.
Having said that, PALM currently doesn’t truly benefit from a cluster: it runs as a single thread without any parallelisation. It may still be useful to run in a cluster environment if you have powerful nodes that have access to the data and can be left running over many days (sometimes none of this is possible in a simple desktop or laptop). Even so, it will use just one machine.
All the best,
Anderson
Hi Anderson,
I want to use PALM software for the statistical test of 2D data which has Ns (number of subjects) x Nm (number of measurement sites) dimensions. I am trying to find which measurement sites are significant.
Some of the elements of the matrix are missing. I see that PALM can handle missing data in the advanced options. However when I use the -imiss option by giving a csv file indicating the missing points in the data as 1s and the rest 0s, I receive an error. Should I use the -dmiss option together with -imiss ?
However, I want to use the same design matrix for all Nm ‘s and only some measurement sites do not have data from let’s say 2 subjects.
I attached my design matrix as an example.
https://drive.google.com/file/d/1jQkFEerl6Blbmqbb91P3-Pyh7-NWUQ6d/view?usp=sharing
Thank you,
Best wishes,
Esin
Hi Esin,
The missing data options in PALM aren’t yet documented because these haven’t been validated. I don’t recommend you use it yet…
Thanks!
Anderson