Consider a set of 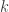 independent tests, each of these to test a certain null hypothesis
independent tests, each of these to test a certain null hypothesis  ,
, 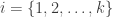 . For each test, a significance level
. For each test, a significance level  , i.e., a p-value, is obtained. All these p-values can be combined into a joint test whether there is a global effect, i.e., if a global null hypothesis
, i.e., a p-value, is obtained. All these p-values can be combined into a joint test whether there is a global effect, i.e., if a global null hypothesis 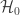 can be rejected.
can be rejected.
There are a number of ways to combine these independent, partial tests. The Fisher method is one of these, and is perhaps the most famous and most widely used. The test was presented in Fisher’s now classical book, Statistical Methods for Research Workers, and was described rather succinctly:
When a number of quite independent tests of significance have been made, it sometimes happens that although few or none can be claimed individually as significant, yet the aggregate gives an impression that the probabilities are on the whole lower than would often have been obtained by chance. It is sometimes desired, taking account only of these probabilities, and not of the detailed composition of the data from which they are derived, which may be of very different kinds, to obtain a single test of the significance of the aggregate, based on the product of the probabilities individually observed.
The circumstance that the sum of a number of values of 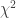 is itself distributed in the
is itself distributed in the 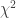 distribution with the appropriate number of degrees of freedom, may be made the basis of such a test. For in the particular case when
distribution with the appropriate number of degrees of freedom, may be made the basis of such a test. For in the particular case when 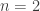 , the natural logarithm of the probability is equal to
, the natural logarithm of the probability is equal to 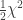 . If therefore we take the natural logarithm of a probability, change its sign and double it, we have the equivalent value of
. If therefore we take the natural logarithm of a probability, change its sign and double it, we have the equivalent value of 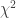 for 2 degrees of freedom. Any number of such values may be added together, to give a composite test, using the Table of
for 2 degrees of freedom. Any number of such values may be added together, to give a composite test, using the Table of 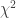 to examine the significance of the result. — Fisher, 1932.
to examine the significance of the result. — Fisher, 1932.
The test is based on the fact that the probability of rejecting the global null hypothesis is related to intersection of the probabilities of each individual test, 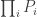 . However,
. However, 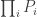 is not uniformly distributed, even if the null is true for all partial tests, and cannot be used itself as the joint significance level for the global test. To remediate this fact, some interesting properties and relationships among distributions of random variables were exploited by Fisher and embodied in the succinct excerpt above. These properties are discussed below.
is not uniformly distributed, even if the null is true for all partial tests, and cannot be used itself as the joint significance level for the global test. To remediate this fact, some interesting properties and relationships among distributions of random variables were exploited by Fisher and embodied in the succinct excerpt above. These properties are discussed below.
The logarithm of uniform is exponential
The cumulative distribution function (cdf) of an exponential distribution is:
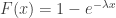
where 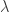 is the rate parameter, the only parameter of this distribution. The inverse cdf is, therefore, given by:
is the rate parameter, the only parameter of this distribution. The inverse cdf is, therefore, given by:

If  is a random variable uniformly distributed in the interval
is a random variable uniformly distributed in the interval ![[0, 1]](https://s0.wp.com/latex.php?latex=%5B0%2C+1%5D&bg=ffffff&fg=333333&s=0&c=20201002) , so is
, so is  , and it is immaterial to differ between them. As a consequence, the previous equation can be equivalently written as:
, and it is immaterial to differ between them. As a consequence, the previous equation can be equivalently written as:
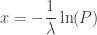
where  , which highlights the fact that the negative of the natural logarithm of a random variable distributed uniformly between 0 and 1 follows an exponential distribution with rate parameter
, which highlights the fact that the negative of the natural logarithm of a random variable distributed uniformly between 0 and 1 follows an exponential distribution with rate parameter  .
.
An exponential with rate 1/2 is chi-squared
The cdf of a chi-squared distribution with  degrees of freedom, i.e.
degrees of freedom, i.e.  , is given by:
, is given by:
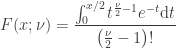
If 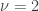 , and solving the integral we have:
, and solving the integral we have:

In other words, a 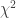 distribution with
distribution with 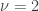 is equivalent to an exponential distribution with rate parameter
is equivalent to an exponential distribution with rate parameter 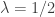 .
.
The sum of chi-squared is also chi-squared
The moment-generating function (mgf) of a sum of independent variables is the product of the mgfs of the respective variables. The mgf of a  is:
is:
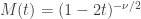
The mgf of the sum of 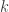 independent variables that follow each a
independent variables that follow each a 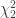 distribution is then given by:
distribution is then given by:
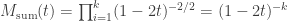
which also defines a 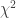 distribution, however with degrees of freedom
distribution, however with degrees of freedom  .
.
Assembling the pieces
With these facts in mind, how to transform the product 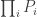 into a p-value that is uniformly distributed when the global null is true? The product can be converted into a sum by taking the logarithm. And as shown above, the logarithm of uniformly distributed variables follows an exponential distribution with rate parameter
into a p-value that is uniformly distributed when the global null is true? The product can be converted into a sum by taking the logarithm. And as shown above, the logarithm of uniformly distributed variables follows an exponential distribution with rate parameter  . Multiplication of each
. Multiplication of each 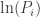 by 2 changes the rate parameter to
by 2 changes the rate parameter to 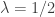 and makes this distribution equivalent to a
and makes this distribution equivalent to a 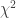 distribution with degrees of freedom
distribution with degrees of freedom 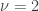 . The sum of
. The sum of 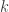 of these logarithms also follow a
of these logarithms also follow a 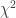 distribution, now with
distribution, now with  degrees of freedom, i.e.,
degrees of freedom, i.e., 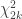 .
.
The statistic for the Fisher method is, therefore, computed as:

with 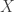 following a
following a 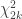 distribution, from which a p-value for the global hypothesis can be easily obtained.
distribution, from which a p-value for the global hypothesis can be easily obtained.
Reference
The details above are not in the book, presumably omitted by Fisher as the knowledge of these derivation details would be of little practical use. Nonetheless, the reference for the book is:
See also
The Fisher’s method to combine p-values is one of the most powerful combining functions that can be used for Non-Parametric Combination.

, where
is the number of images averaged.