
Can you tell?
The now famous story is that in an otherwise unremarkable summer afternoon in Cambridge in the 1920’s, a group of friends eventually discussed about the claims made by one of the presents about her abilities on discriminating whether milk was poured first or last when preparing a cup of tea with milk. One of the presents was Ronald Fisher, and the story, along with a detailed description of how to conduct a simple experiment to test the claimed ability, and how to obtain an exact solution, was presented at length in the Chapter 2 of his book The Design of Experiments, a few lines of which are quoted below:
A lady declares that by tasting a cup of tea made with milk she can discriminate whether the milk or the tea infusion was first added to the cup. We will consider the problem of designing an experiment by means of which this assertion can be tested. […] [It] consists in mixing eight cups of tea, four in one way and four in the other, and presenting them to the subject for judgment in a random order. The subject has been told in advance of that the test will consist, namely, that she will be asked to taste eight cups, that these shall be four of each kind […]. — Fisher, 1935.
There are distinct possible orderings of these cups, and by telling the subject in advance that there are four cups of each type, this guarantees that the answer will include four of each.
The lady in question eventually answered correctly six out of the eight trials. The results can be assembled in a 2 by 2 contingency table:
| True order: | Total (margin) | |||
|---|---|---|---|---|
| Tea first | Milk first | |||
| Lady’s Guesses: | Tea first | |||
| Milk first | ||||
| Total (margin) | ||||
With these results, what should be concluded about the ability of the lady in discriminating whether milk or tea was poured first? It is not possible to prove that she would never be wrong, because if a sufficiently large number of cups of tea were offered, a single failure would disprove such hypothesis. However, a test that she is never right can be disproven, with a certain margin of uncertainty, given the number of cups offered.
Solution using Fisher’s exact method
Fisher presented an exact solution for this experiment. It is exact in the sense that it allows an exact probability to be assigned to each of the possible outcomes. The probability can be calculated as:
For the particular configuration of the contingency table above, the probability is . This is not the final result, though: what matters to disprove the hypothesis that she is not able to discriminate is how likely it would be for her to find a result at least as extreme as the one observed. In this case, there is one case that is more extreme, which would be the one in which she would have made correct guesses for all the 8 cups, in which case the values in the contingency table above would have been
,
,
, and
, with a probability computed with the same formula as
. Adding these two probabilities together yield
.
Thus, if the lady is not able to discriminate whether tea or milk was poured first, the chance of observing a result at least as favourable towards her claim would be 0.24286, i.e., about 24%.
If from the outset we were willing to consider a significance level 0.05 (5%) as an informal rule to disprove the null hypothesis, we would have considered the p-value = 0.24286 as non-significant. This p-value is exact, a point that will become more clear below, in the section about permutation tests.
Using the hypergeometric distribution directly
The above process can become laborious for experiments with larger number of trials. An alternative, but equivalent solution, is to appeal directly to the hypergeometric distribution. The probability mass function (pmf) of this distribution can be written as a function the parameters of the contingency table as:
The pmf is equivalent to Fisher’s exact formula to compute the probability of a particular configuration. The cumulative density function, which is conditional on the margins being fixed, is:
where . Computing from the above (details omitted), yield the same value as using Fisher’s presentation, that is, the p-value is (exactly) 0.24286.
Solution using Pearson’s 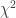 method
method
Much earlier than the tea situation described above, Karl Pearson had already considered the problem of inference in contingency tables, having proposed a test based on a statistic.
where is the observed value for the element in the position
in the table,
and
are respectively the number of rows and columns, and
is the expected value for these elements if the null hypothesis is true. The values
can be computed as the product of the marginals for row
and column
, divided by the overall number of observations
. A simpler, equivalent formula is given by:
A p-value can be computed from the distribution with degrees of freedom
.
Under the null, we can expect a value equal to 2 in each of the 2 cells, that is, the lady would for each cup have a 50:50 chance of answering correctly. For the original tea tasting experiment, Pearson’s method give quite inaccurate results: , which corresponds to a p-value of 0.07865. However, it is well known that this method is inaccurate if cells in the table have too small quantities, usually below 5 or 6.
Improvement using Yates’ continuity correction
To solve the above well-known issue with small quantities, Yates (1934) proposed a correction, such that the test statistic becomes:
Applying this correction to the original tea experiment gives , and a p-value of 0.23975, which is very similar to the one given by the Fisher method. Note again that this approach, like the
test, predates Fisher’s exact test.
Equivalence of Fisher’s exact test and permutation tests
The method proposed by Fisher corresponds to a permutation test. Let be a column vector containing binary indicators for whether milk was truly poured first. Let
be a column vector containing binary indicators for whether the lady answered that milk was poured first. The general linear model (GLM) can be used as usual, such that
, where
is a regression coefficient, and
are the residuals.
Under the null hypothesis that the lady cannot discriminate, the binary values in can be permuted randomly. There are
possible unique rearrangements. Out of these, in 17, there are 6 or more (out of 8) correct answers matching the values in
, which gives a p-value 17/70 = 0.24286.
Note that the strategy using the GLM can be used even if both variables and
are binary, as in the example of the tea tasting, even if the residuals are not normally distributed (permutation tests do not depend on distributional assumptions), and even considering that values in
can lead to non-sensical predictions in
, as prediction is not the objective of this analysis.
Why not a binomial test?
The binomial test could be considered if the lady did not know in advance that there were 4 cups of each mixture order. Since she knew (so the two margins of the table are fixed), each cup was not independent from each other, and her possible answers had to be constrained by answers previously given. The binomial test assumes independence, thus, is not an option for this analysis.
Relevance
Using this simple experiment, Fisher established most of the fundamental principles for hypothesis testing, which contributed to major advances across biological and physical sciences. A careful read of the original text shows a precise use of terms, in a concise and unambiguous presentation, in contrast with many later and more verbose textbooks that eventually hid from readers most of the fundamental principles.
References
- Fisher, R. A. The Design of Experiments. Oliver and Boyd, Edinburgh, 1935.
- Pearson K. X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philos Mag Ser 5. 1900 Jul;50(302):157–75.
- Yates F. Contingency Tables Involving Small Numbers and the Chi^2 test. Suppl to J R Stat Soc. 1934;1(2):217–35.
The photograph at the top (tea with milk) is in public domain.


